| miR-451 | Regulating normal erythropoiesis; downregulated in beta-thalassemia leading to erythroid hyperplasia |
| miR-24-1 | Part of miR-23b/27b/24-1 cluster suppress fibrotic of liver, disturbing hepcidin |
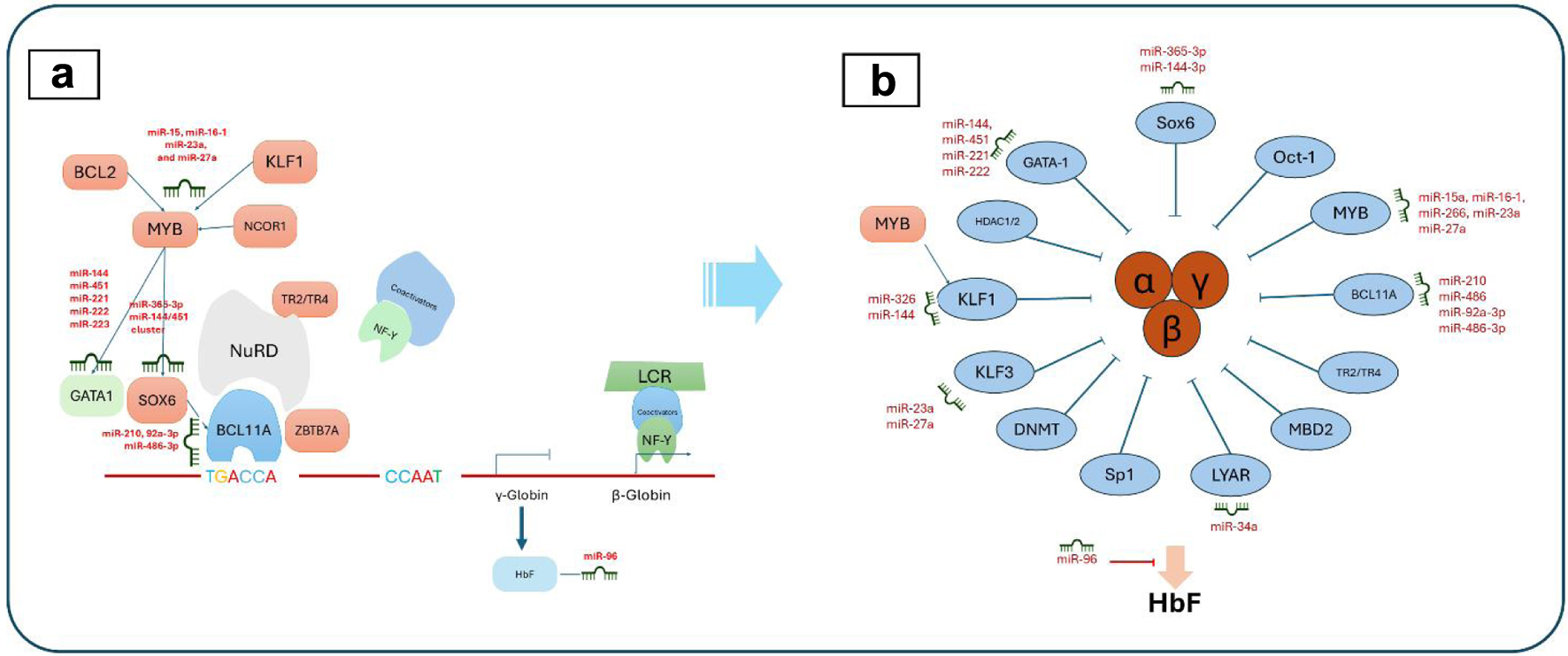
| miR-92a-3p | Regulating gamma-globin, GSH, SOD, ROS, and cell apoptosis |
| miR-16-5p | Associated with hemolysis/transfusion reactions |
| miR-210 | Suppressing erythropoiesis in alpha-thalassemia; modulating oxidative stress |
| miR-let-7b | Controlling inflammation by regulating the NF-κB pathway |
| miR-20a | Regulating iron metabolism by targeting ferritin; potential biomarker for iron overload and liver damage |
| miR-144 | Increased sensitivity to oxidative stress |
| miR-221 | Erythroid cell apoptosis |
| miR-let-7b | Regulating ferroportin and iron metabolism |
| miR-155 | Modulating immune reactions associated with transfusions |
| miR-21 | Potential biomarker for iron overload and liver damage |
| miR-155 | Controlling inflammation by regulating the NF-κB pathway |
| miR-222 | Upregulated in alpha-thalassemia leading to erythroid cell apoptosis |
| miR-92a-3p | Associated with hemolysis/transfusion reactions |
| miR-27b | Part of miR-23b/27b/24-1 cluster suppress fibrotic of liver |
| miR-9 | Suppressing FoxO3 and affecting ROS production |
| miR-485-3p | Regulating iron metabolism by targeting TfR1 |
| miR-214 | Upregulated in thalassemia patients with oxidative stress |
| miR-146a | Potential biomarker for inflammation-related complications like sepsis |
| miR-122 | Regulating hepcidin and iron metabolism |
| miR-16 | Regulating normal erythropoiesis; downregulated in beta-thalassemia leading to erythroid hyperplasia |
| miR-150 | Inhibiting proliferation and differentiation of erythroid progenitor cells; modulating immune cell function (T and B lymphocytes); potential biomarker for erythropoiesis |