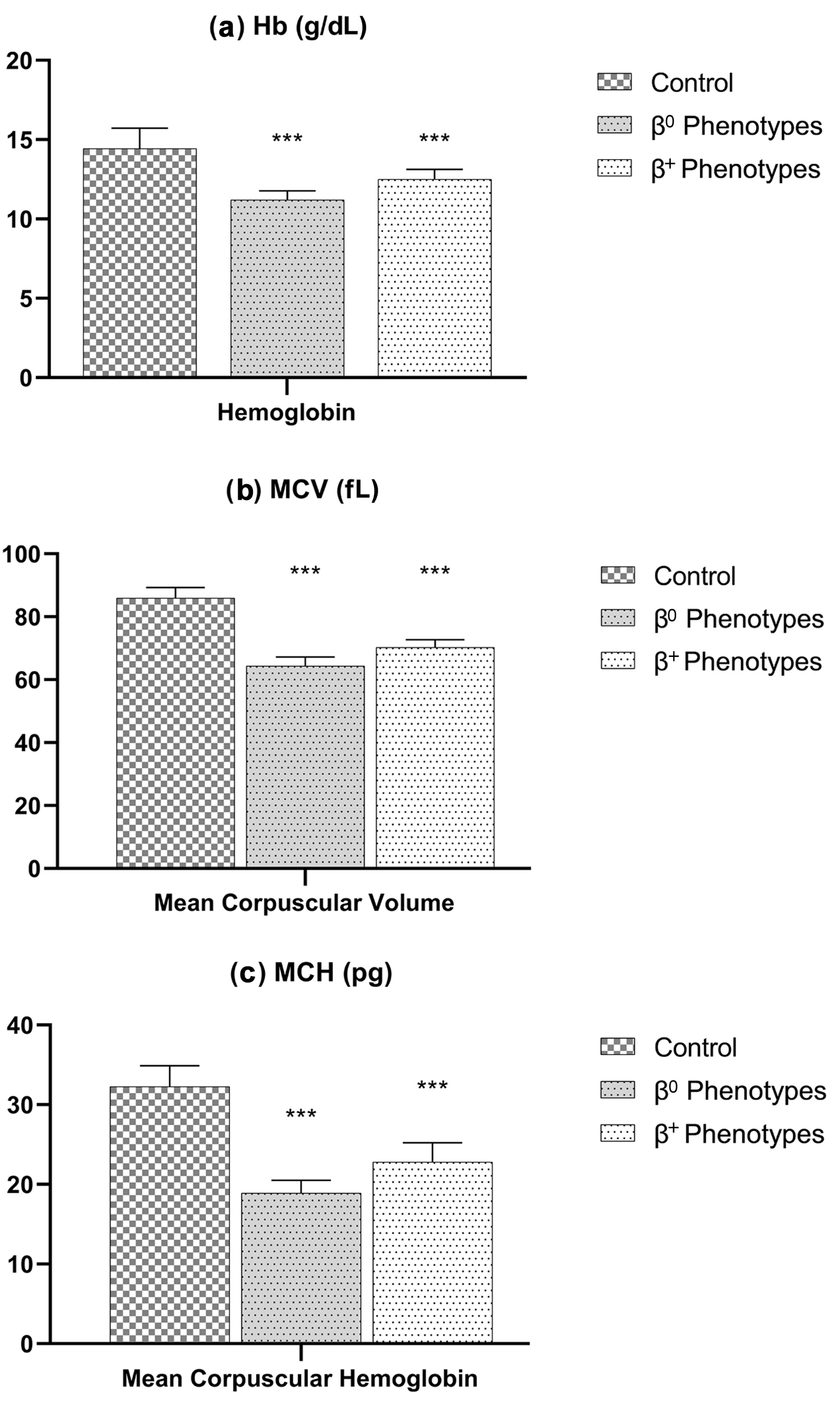
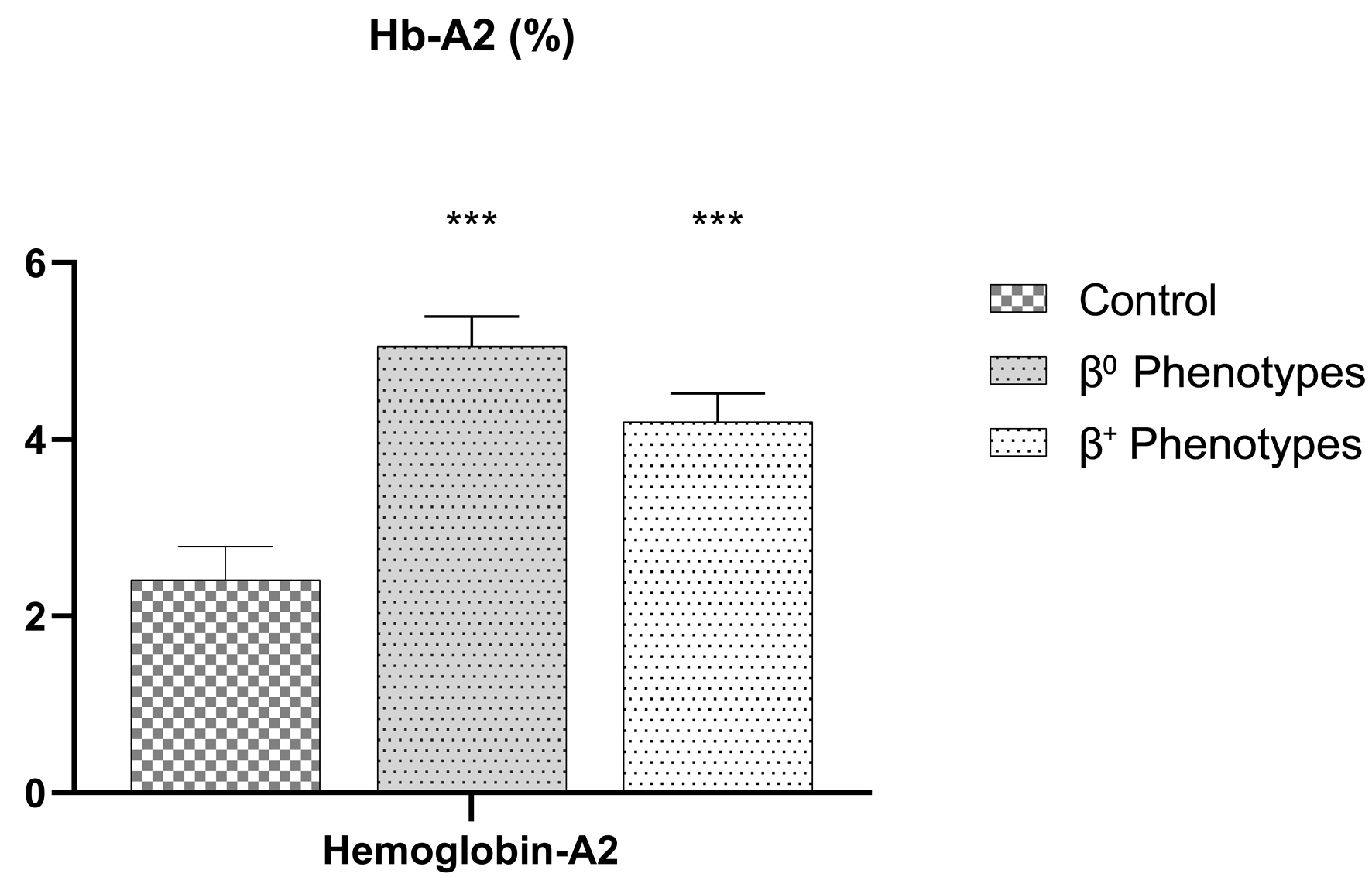
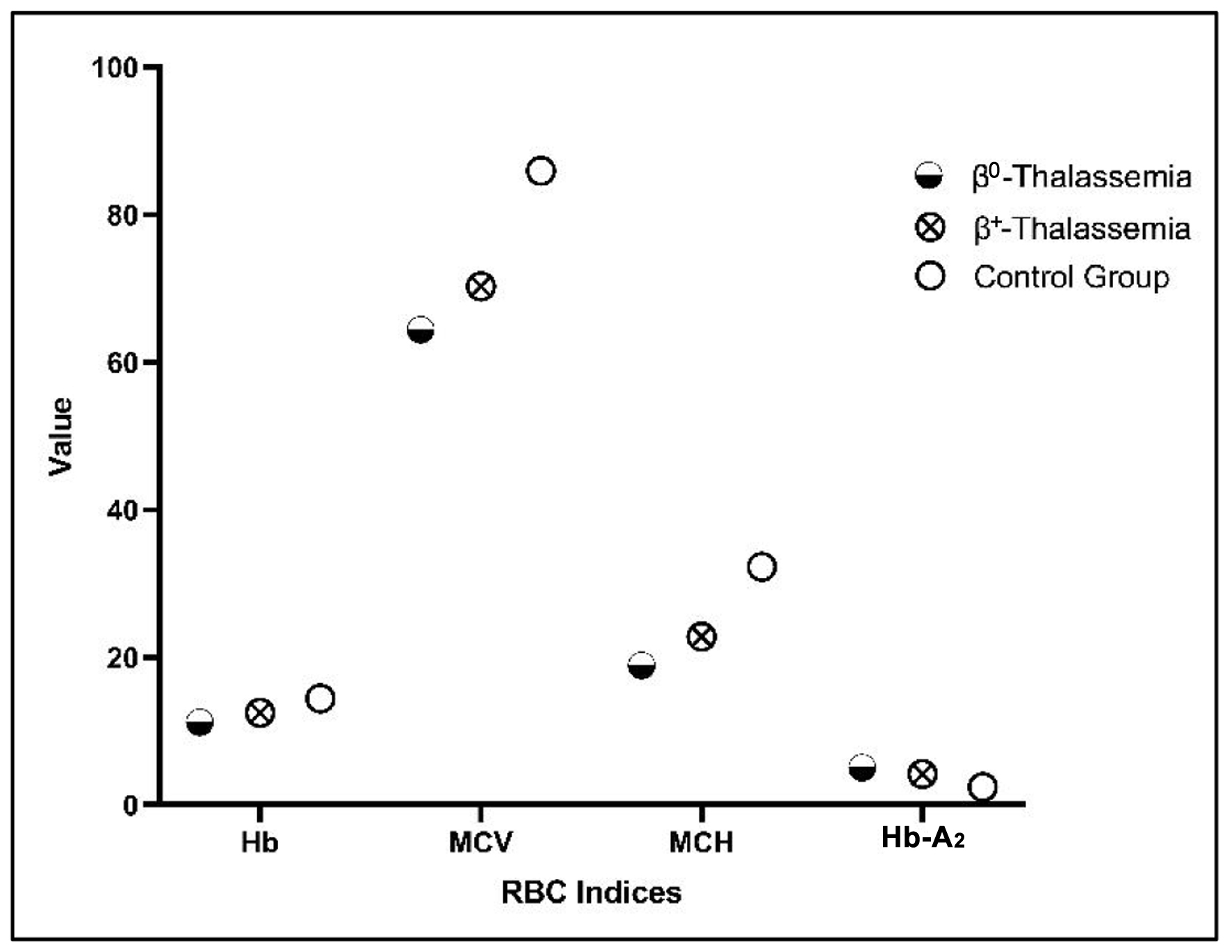
| β0 phenotypes (N = 64) (44.1%) | | | | | | |
| IVS 2.1 [G>A] | HBB: c.315+1G>A | RNA processing/splice junction | β0 | Mediterranean, US Blacks | 21 | 32.8 |
| IVS 1.1 [G>A] | HBB: c.92+1G>A | RNA processing/splice junction | β0 | Mediterranean | 18 | 28.1 |
| Codon 8 [-AA] | HBB: c.25_26delAA | RNA translation/frameshift | β0 | Mediterranean | 8 | 12.5 |
| Codon 5 [-CT] | HBB: c.17_18delCT | RNA translation/frameshift | β0 | Mediterranean | 7 | 10.9 |
| Codon 39 [C>T] | HBB: c.118C>T | RNA translation/nonsense codons | β0 | Mediterranean | 7 | 10.9 |
| Codon 6 [-A] | HBB: c.20delA | RNA translation/frameshift | β0 | Mediterranean, US Blacks | 1 | 1.6 |
| IVS 1.130 [G>C] | HBB: c.93-1G>C | RNA processing/splice junction | β0 | Italian, Japanese, UAE | 1 | 1.6 |
| Codon 44 [-C] | HBB: c.135delC | RNA translation/frameshift | β0 | Kurdish | 1 | 1.6 |
| β+ phenotypes (N = 81) (55.9%) | | | | | | |
| IVS 1.6 [T>C] | HBB: c.92+6T>C | RNA processing/consensus splice sites | β+ | Mediterranean | 28 | 34.6 |
| IVS 1.110 [G>A] | HBB: c.93-21G>A | RNA processing/cryptic splice sites | β+ | Mediterranean | 21 | 25.9 |
| IVS 2.745 [C>G] | HBB: c.316-106C>G | RNA processing/cryptic splice sites | β+ | Mediterranean | 16 | 19.8 |
| IVS 1.5 [G>C] | HBB: c.92+5G>C | RNA processing/consensus splice sites | β+ | Asian Indian, SE Asian | 5 | 6.2 |
| -30 [T>A] | HBB: c.-80T>A | Transcriptional variants/promoter regulatory elements | β+ | Mediterranean, Bulgarian | 4 | 4.9 |
| -87 [C>G] | HBB: c.-137C>G | Transcriptional variants/promoter regulatory elements | β++ | Mediterranean | 3 | 3.7 |
| -101 [C>T] | HBB: c.-151C>T | Transcriptional variants/promoter regulatory elements | β++ (silent) | Mediterranean | 3 | 3.7 |
| IVS 2.848 [C>A] | HBB: c.316-3C>A | RNA processing/consensus splice sites | β+ | UB Blacks, Egyptian, Iranian | 1 | 1.2 |