| Journal of Clinical Medicine Research, ISSN 1918-3003 print, 1918-3011 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Clin Med Res and Elmer Press Inc |
| Journal website https://jocmr.elmerjournals.com |
Original Article
Volume 16, Number 7-8, August 2024, pages 355-362
Renin-Angiotensin System Genes Polymorphisms in Patients With COVID-19 and Its Relation to Severe Cases of SARS-CoV-2 Infection
Anna E. Braginaa, b, e , Aida I. Tarzimanovaa
, Yulia N. Rodionovaa, b
, Ekaterina S. Ogibeninac
, Aleksandr Yu. Suvorovb
, Natalya A. Druzhininaa, b
, Lyubov V. Vasilyevaa
, Tatiana I. Ishinaa
, Ivan D. Medvedeva
, Marina S. Borlakovad
, Anastasiia R. Komelkovaa
, Daria V. Gushchinaa
, Artem A. Khachaturova
, Valery I. Podzolkova
a2nd Internal Medicine (2nd Faculty Therapy) Department, N.V. Sklifosovskiy Institute of Clinical Medicine, Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
bWorld-Class Research Center “Digital biodesign and Personalized Healthcare”, Sechenov First Moscow State Medical University, Moscow, Russia
cUniversity Clinical Hospital #4, I.M. Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
dDepartment of Pharmacology, Institute of Digital Biodesign and Modeling of Living Systems, Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
eCorresponding Author: Anna E. Bragina, 2nd Internal Medicine (2nd Faculty Therapy) Department, N.V. Sklifosovskiy Institute of Clinical Medicine, Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
Manuscript submitted June 4, 2024, accepted July 6, 2024, published online July 18, 2024
Short title: RAS Genes Polymorphisms in Patients With COVID-19
doi: https://doi.org/10.14740/jocmr5223
| Abstract | ▴Top |
Background: Different variants of single nucleotide polymorphisms (SNPs) of angiotensinogen (AGT), angiotensin-converting enzyme type 1 (ACE1), and angiotensin II receptors type 1 (AGTR1) and 2 (AGTR2) genes determine different susceptibility to cardiovascular disease (CVD) and hypertension, which can be considered as risk factors for fatal outcomes among coronavirus disease 2019 (COVID-19) patients. The objective of our study was to assess the relation between the frequency of SNPs of the renin-angiotensin system (RAS) components, and the severity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
Methods: The cross-sectional study included 100 patients with a laboratory-confirmed diagnosis of COVID-19 admitted to the hospital. Criteria for severe COVID-19 included respiratory rate (RR) > 30/min, blood oxygen saturation (SpO2) ≤ 93%, signs of unstable hemodynamics with systolic blood pressure (SBP) < 90 and/or diastolic blood pressure (DBP) < 60 mm Hg. All patients were identified with alleles and genotypes of the polymorphic markers rs4762 of the AGT gene, rs1799752 of the ACE1 gene, rs5186 of the AGTR1 gene and rs1403543 of the AGTR2 gene using the polymerase chain reaction method in human DNA preparations on real-time CFX96C1000 Touch, Bio-Rad equipment (Syntol, Russia). Statistical analysis was performed in R v.4.2.
Results: Patients were divided into groups with severe (n = 44) and moderate COVID-19 (n = 56). For ACE1 rs1799752, a significant deviation from the population distribution was detected in both studied subgroups. A higher frequency of the C allele SNP rs5186 AGTR1 gene was detected in the group with severe disease. More frequent A/A genotype of SNP rs1403543 AGTR2 was detected among females with severe COVID-19. Haplotype analysis revealed more common DCG haplotype among patients with severe COVID-19. The odds ratio for severe COVID-19 in the presence of the DCG haplotype was 3.996 (95% confidential interval: 1.080 -14.791, P < 0.05).
Conclusions: Our data suggest that the SNP genes of the RAS components, may allow to identify groups of patients predisposed to a more severe course of COVID-19.
Keywords: COVID-19; SARS-CoV-2 infection; Single nucleotide polymorphisms; Angiotensinogen gene; Angiotensin-converting enzyme type 1 gene; Angiotensin II receptor type 1 gene; Angiotensin II receptor type 2 gene
| Introduction | ▴Top |
Disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to the 2020 - 2022 coronavirus pandemic with high morbidity and mortality, introduction of total quarantine measures, social and economic collapse. Despite the official announcement by the World Health Organization of the end of the global pandemic, the morbidity of coronavirus disease 2019 (COVID-19) remains at a high level. The relation between the cardiovascular risk factors, cardiovascular diseases (CVDs), and all-cause in-hospital mortality in patients with COVID-19 is well established [1-3]. Thus, according to meta-analyses made by Dessie et al [4] and Degarege et al [5], CVD, diabetes, and hypertension can be considered as risk factors for fatal outcomes among COVID-19 patients.
The relevance of the association between COVID-19 and CVDs might be explained by the pathogenesis of the coronavirus infection. The key factor for SARS-CoV-2 entering the cells is angiotensin-converting enzyme type 2 (ACE2) with participation of transmembrane serine proteases type II. ACE2 is a transmembrane protein involved in the breakdown of angiotensin II (1-8) to form angiotensin (1-7). It has the properties of a vasodilator and a functional angiotensin II antagonist [6]. A decrease in ACE2 activity due to various reasons leads to the activation of other components of the renin-angiotensin system (RAS) with all its pathologic consequences such as activation of inflammation, thrombogenic potential, and oxidative processes [6].
The genetic heterogeneity of RAS components is well known. Different variants of single nucleotide polymorphisms (SNPs) of angiotensinogen (AGT), angiotensin-converting enzyme type 1 (ACE1), and angiotensin II receptor types 1 and 2 (AGTR1 and AGTR2) genes determine different susceptibility to CVDs and hypertension. The most studied is I/D polymorphism in ACE1 rs1799752, for which a significant association with the prevalence and severity of the course of CVDs has been shown [7]. There are data on the association of T/T and C/T genotypes of the AGT rs4762 gene [8, 9], C/C and T/T genotypes of the AGTR1 rs5186 gene [10, 11] with hypertension and preeclampsia. There are a few studies of RAS gene polymorphism in symptomatic patients with COVID-19 [12, 13] and in COVID-19-related retinopathy [14].
This work aims to assess the relation between the frequency of SNPs of the AGT, ACE1 genes, and AGTR1 and AGTR2, and the severity of SARS-CoV-2 infection.
| Materials and Methods | ▴Top |
The cross-sectional study included 100 Caucasian patients who were treated at the University Clinical Hospital No. 4 of Sechenov University with a laboratory-confirmed diagnosis of COVID-19 from November 2021 to February 2022. Written informed consent was obtained from all patients voluntarily joined the study. Research protocol was approved by the Locale Ethical Committee of Sechenov University. The study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
Inclusion criteria for the study were patient age > 18 years and informed consent for the study. Exclusion criteria: acute coronary syndrome, acute cerebrovascular accident, pulmonary embolism, severe liver or kidney pathology (glomerular filtration rate (GFR) < 15 mL/min/1.73 m2), hyperkalemia > 5.0 mmol/L, severe anemia, cancer. Patients underwent standard laboratory and instrumental examinations [15]. All patients were symptomatic.
The data on the severity of SARS-CoV-2 infection were taken from the discharge summaries. The severity of the coronavirus infection was assessed in accordance with the temporary guidelines of the Russian Ministry of Health. Criteria for severe COVID-19 included respiratory rate (RR) > 30/min, blood oxygen saturation (SpO2) ≤ 93%, signs of unstable hemodynamics with systolic blood pressure (SBP) < 90 and/or diastolic blood pressure (DBP) < 60 mm Hg [15], which meet international criteria for the severity of COVID-19 [16]. The extent of pulmonary impairment was evaluated based on the established categorization according to the results of multi-slice spiral computed tomography (CT) of the chest: CT0 denotes the nonexistence of inflammation foci and infiltrates; CT1 signifies the manifestation of viral pneumonia symptoms, encompassing up to a quarter of the pulmonary tissue; CT2 corresponds to the breadth of lung impairment ranging from 25% to 50%; CT3 relates to the pulmonary tissue damage spanning from 50% to 75%; CT4 represents injury to over 75% of the pulmonary tissue.
All patients (n = 100) were identified with alleles and genotypes of the polymorphic markers rs4762 of the AGT gene, rs1799752 of the ACE1 gene, rs5186 of the AGTR1 and rs1403543 of the AGTR2 using the polymerase chain reaction method in the real-time in human deoxyribonucleic acid (DNA) preparations. They were obtained from venous blood using allele-specific TagMan probes on real-time CFX96C1000 Touch, Bio-Rad equipment. To isolate DNA from the analyzed material, a set of reagents DNA-EXTRAN-1 (Syntol, Russia) was used. As a result, alleles C and T, genotypes C/C, T/T, C/T were identified for the rs4762 polymorphic marker of the AGT gene; alleles I and D, genotypes I/I, D/D, D/I were identified for the polymorphic marker rs1799752 of the ACE1 gene; alleles A and C, genotypes A/A, C/C, A/C were identified for the polymorphic marker rs5186 of the AGTR1 gene; alleles G and A, genotypes G/G, A/A, G/A were identified for the polymorphic marker rs1403543 of the AGTR2 gene.
Statistical analysis was performed in R v.4.2. For categorical data, the proportions and absolute values were determined. Comparative analysis of quantitative variables was carried out using the Mann-Whitney U test, categorical and qualitative features using Pearson’s Chi-square criterion, if not applicable, Fisher’s exact test was used. The odds ratio (OR) was calculated to assess the link between genotypes and COVID-19 severity. The frequencies of SNPs rs1799752, rs5186, and rs1403543 were compared with the population prevalence of these genomes and alleles obtained in the study by Kuznetsova et al, 2004 about the distribution of genetic variants of the RAS genes in the East Slavic population [17]. Allele frequencies of SNPs rs4762 were compared with their prevalence in the European population obtained from the open database 1000 Genomes Browsers (A Deep Catalog of Human Genetic Variation) [18, 19]. Genome association analysis was performed using the SNPassoc library [20] and haplo.stats library [21]. For genotypes, the significance of the Hardy-Weinberg equilibrium (HWE) was calculated. Statistics were calculated for the whole group, depending on the severity and gender. Haplotype analysis included the estimation of frequencies in the whole sample and by severity (moderate or severe). Scores and significance were adjusted for age and gender.
| Results | ▴Top |
According to the severity criteria of COVID-19 (RR, SpO2 and hemodynamic instability), patients were divided into groups with severe (44 people) and moderate coronavirus infection (56 people). The main clinical and demographic characteristics of the groups are given in Table 1. Age, gender, levels of SBP, DBP, and heart rate (HR) of the groups did not differ. There were substantial differences between RR and SpO2, that were explained by the group selection criteria. Along with this, patients with severe COVID-19 had higher concentration of C-reactive protein (CRP), ferritin, D-dimer, and CT-grade of pulmonary injury. The analysis of concomitant pathology did not reveal any significant differences in the frequency of comorbidities. Half of the patients, included in the study, took antihypertensives. Thirty-three percent (33%) of the whole group received anti-RAS drugs (either ACE inhibitors or angiotensin II receptor blockers): 34.1% in group with severe COVID-19 vs. 32.1% in moderate severe COVID-19 (P = 0.764).
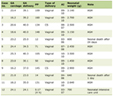 Click to view | Table 1. Main Clinical and Demographic Characteristics of Patients With COVID-19 |
The distribution of various genotypes and alleles frequency of the studied genes in patients with severe and moderate COVID-19 is presented in Table 2. The subgroups revealed the equal distribution of genotypes and alleles of SNP AGT rs4762 without deviation from the HWE. For SNP ACE1 rs1799752, a significant deviation from the HWE was detected in both studied subgroups. A predominance of D alleles of ACE1 polymorphism rs1799752 was revealed compared to the population data (D-allele 52.9%, I-allele 47.1% [16]): χ2 = 17.73, P < 0.001 for severe COVID-19 and χ2 = 25.69, P < 0.001 for moderate COVID-19. Groups with severe and moderate disease did not differ significantly in allele frequency of ACE1 rs1799752. Significant differences in the distribution of alleles and genotypes depending on the severity of COVID-19 were for SNP rs5186 of the AGTR1 gene. A higher frequency of the C allele was detected in the group with severe disease. The AGTR2 rs1403543 gene is in the X chromosome, and therefore the distributions of genotypes and alleles of this SNP were analyzed separately in males and females. A significant deviation in the distribution of SNP rs1403543 from the HWE was revealed in a male subgroup with both severe and moderate disease. In the female group with different severity of COVID-19, there was a significant difference in the genotype frequencies of SNP rs1403543 (Table 2).
 Click to view | Table 2. Frequency Distribution of Genotypes and Alleles of Studied Polymorphisms in Patients With Severe and Moderate COVID-19 |
Haplotype distribution analysis was performed using rs1799752, rs5186, and rs1403543 SNPs that demonstrated deviations from HWE, population distribution, and/or intergroup differences. Thus, nine variants of haplotypes were formed, and their frequencies are presented in Table 3. The DCG haplotype was significantly more common among patients with severe COVID-19. The distribution of other haplotypes did not differ significantly between subgroups. The ICG haplotype was the rarest and was not detected in patients with severe COVID-19 in the examined cohort. The OR for severe COVID-19 in the presence of the DCG haplotype was 3.996 (95% confidence interval (CI): 1.080 - 14.791, P < 0.05).
 Click to view | Table 3. Haplotype Analysis Performed on ACE1 rs1799752, AGTR1 rs5186, and AGTR2 rs1403543 SNPs and Their Corresponding Frequencies in Moderate (N = 56) and Severe (N = 44) COVID-19 |
| Discussion | ▴Top |
We studied the SNP of AGT rs4762, ACE1 rs1799752, AGTR1 rs5186 and AGTR2 rs1403543 genes in a cohort of 100 patients hospitalized with confirmed COVID-19. To identify relation between the frequency of SNP and the severity of COVID-19, we have formed two groups, depending on the severity of the disease: 44 patients with severe and 56 patients with moderate COVID-19.
We have shown a significantly higher frequency of I/D, D/D genotypes and the D-allele of the ACE1 rs1799752 gene among patients hospitalized with COVID-19 compared to the Russian population [17]. The ACE1 gene is in chromosome 17 (17q23.3). The nature of the ACE1 rs1799752 gene polymorphism is in the insertion (insertion, I) or loss (deletion, D) of the Alu repeat in 289 nucleotide pairs in the 16th intron. Deletion of the Alu repeat is accompanied by a significant increase in the expression of the ACE1 gene and a rise of ACE1 levels [22]. The increase in ACE1 levels occurs even in case of heterozygous status (I/D). The highest level is observed in patients with homozygous genotype D/D rs1799752, which is twice higher than in patients with genotype I/I. The relation between the D/D genotype and a wide range of CVDs, including coronary heart disease (CHD), heart attack, left ventricular hypertrophy, hypertension, kidney disease, and neurodegenerative diseases has been detected [23].
The second gene involved in the formation of RAS activity and demonstrated significant differences in the distribution of its polymorphic variants was the AGTR1 A1166C A>C gene (rs5186). The frequency of the C allele in our cohort was significantly higher in patients with severe COVID-19 - 29.6% vs. 17% in the group of moderate disease. The AGTR1 gene is in chromosome 3 (3q24) and encodes type 1 receptors for AT-II, located in the vascular endothelium and mediating the majority of angiotensin II cardiovascular effects. More than 50 polymorphic variants of the AGTR1 gene are known. The A1166C A>C (rs5186) polymorphism, which was studied in our work, has the greatest clinical significance. It is characterized by the replacement of the nucleotide adenine with cytosine at position 1166 of DNA. The presence of the mutant allele C in this polymorphism leads to increased sensitivity of AGTR1 to normal levels of angiotensin II and, consequently, to higher BP levels. Studies have shown that hypertensives are significantly more common to have the A/C or C/C genotype of the AGTR1 gene compared to normotensives [24].
We have studied two more SNPs that are important for the functioning of the RAS: the AGT gene and AGTR2. The AGT gene is located on chromosome 1 (1q42). The replacement from cytosine (C) to thymine (T) in 521 positions leads to a change in the amino acid sequence of the protein structure of AGT with the replacement from tryptophan to methionine in 174 positions, which is designated as T207M C>T (rs4762). It has been shown that the presence of the T-allele is associated with a higher level of AGT expression and the development of hypertension [25]. In groups of older people with hypertension, the C/T genotype occurs about five times more often than in the control group without CVDs. Also, the presence of the T allele in the genotype increases the risk of developing cardiometabolic disorders and CHD [8, 9]. In our study, the frequencies of SNP AGT rs4762 have corresponded to the HWE and have not differed between groups of hospitalized patients with severe and moderate COVID-19.
SNP AGTR2 rs1403543 is characterized by a replacement from guanine (G) to adenine (A) at nucleotide position 1675 of the genome (genetic marker G1675A rs1403543) and is associated with a change in gene expression regulation. The G allele is associated with the activation of transcription and the increase in the number of AGTR2 on the cell membrane. This leads to effects opposite to those caused by angiotensin II, such as vasodilation, natriuresis, and slowing of smooth muscle hypertrophy. Carriers of the A allele have an increased risk of hypertension, pregnancy complications, and CHD, which is caused by a decrease in AGTR2 expression and a shift in the functional activity of the RAS towards AGTR1 [26]. The AGTR2 gene is in the X chromosome, that’s why the distribution of its polymorphic variants has gender specificity. In our study, the frequencies of SNP AGTR2 rs1403543 alleles have deviated from the HWE in males with severe and moderate COVID-19. In females with different COVID-19 severity, a significant difference in the distribution of rs1403543 genotypes was revealed (χ2 = 10.570, P = 0.006).
The influence of polymorphism of the RAS genes on coronavirus infection has been studied in several research. The study by Kouhpayeh et al 2021 revealed differences in the frequencies of SNP genes in patients with COVID-19 and in the control group, which the authors explained by the different prevalence of CVDs, that might be caused by certain AGT, ACE1, AGTR1 genes polymorphisms of the [13]. However, in our study, in the subgroups with severe and moderate COVID-19, the frequency of comorbidities such as hypertension, CHD, diabetes, CKD, and chronic heart failure (CHF) has not differed.
The hypothesis of the role of the ACE 1 rs1799752 polymorphism in the pathogenesis of COVID-19 was formulated in 2020 by Gemmati et al [27]. The authors showed that more severe course of the disease and higher mortality rates among patients of the Caucasian race were correlated with D/D polymorphism in the ACE1 gene. This phenomenon was associated with hyperactivation of the RAS. However, there was no association of ACE1 rs1799752 polymorphism with the severity of COVID-19 infection in a systematic review and meta-analysis published in 2022 by Gupta et al [28]. However, this meta-analysis has significant limitations, as it compares only severe vs. non-severe COVID-19, without asymptomatic and control groups.
We also did not reveal differences in the frequency of ACE1 I/D polymorphism between the groups of severe and moderate COVID-19. However, an important result of our study was the deviation of the frequency distribution of ACE1 rs1799752 genotypes from the HWE and of population data in the whole group and each of the comparison groups. This fact may be related to the higher prevalence of the D-allele of the ACE1 gene among patients with COVID-19. The results of a study by Yamamoto et al about the negative correlation of the ACE1 I/I genotype with susceptibility to SARS-CoV-2 infection confirm this [29]. Cafiero et al [12] obtained similar data on a significantly higher frequency of I/D and D/D genotypes among symptomatic COVID-19 patients vs. asymptomatic.
The association of AGTR1 rs5186 polymorphism with COVID-19 has been studied by few scientific groups. Gupta and co-authors showed a reliable increase in the risk of severe COVID-19 in C-allele vs. A-allele (OR: 1.49, 95% CI: 1.13 - 1.98, P = 0.005) [28]. Our results are similar. The value of the AGTR2 rs1403543 polymorphism in COVID-19 has not been widely studied. The borderline relation between the AA genotype of the SNP AGTR2 rs1403543 and the presence of COVID-associated retinopathy in males (P = 0.05) was found in the only study by Jevnikar et al 2022 [14].
We analyzed the combination of SNP alleles of the RAS genes in our research and found that the DCG haplotype is more common in patients with severe COVID-19. It means that this haplotype increases the risk of severe COVID-19 with OR 3.996. Based on these data, this allelic combination can be used as a predictor of more severe forms of COVID-19.
The relation between RAS activity and COVID-19 severity seems to be mutual. On the one hand, decrease of ACE2 level due to blocking by viral particles, reduction in expression, and splitting by transmembrane metalloprotease 17 can contribute to reduced transformation of angiotensin I and II into angiotensin 1-9 and 1-7, respectively. Thus, it might lead to a shift of the RAS activity to activation of angiotensin II synthesis [30]. On the other hand, angiotensin II by acting on AGTR1 can significantly increase ACE2 level by mRNA and therefore potentiate SARS-CoV-2 activity, as evidenced by the data of Caputo et al [31]. Thus, high AGTR1 expression with the C-allele of rs5186 polymorphism, especially in high ACE1 concentrations in patients with D-allele predominance of the ACE1 rs1799752 gene, can significantly increase RAS activity during the active phase of COVID-19.
Our study showed significant shifts in the allelic distribution of the SNP genes ACE1 rs1799752, AGTR1 rs5186 and AGTR2 rs1403543 in the cohort of hospitalized patients with COVID-19. We have identified the DCG haplotype which carries a higher risk of severe COVID-19. A relatively small sample, the absence of outpatients with mild COVID-19 and a control group of patients without COVID-19 can be considered as limitations of our work, that is why additional research in this area remains feasible.
Conclusion
The modern paradigm of medicine, based on a personalized approach and predictive strategy, requires the search for genetic markers of socially significant diseases. Our data suggest that the SNP genes of the RAS components, namely ACE1 rs1799752, AGTR1 rs5186, and AGTR2 rs1403543, may allow us to identify groups of patients predisposed to a more severe course of COVID-19. Certainly, testing this hypothesis requires additional studies with larger samples and comparison groups from non-hospitalized patients with mild and asymptomatic forms of the disease.
Acknowledgments
None to declare.
Financial Disclosure
This work was financed by the Ministry of Science and Higher Education of the Russian Federation within the framework of state support for the creation and development of World-Class Research Centers “Digital Biodesign and Personalized Healthcare” No. 075-15-2022-304.
Conflict of Interest
The authors confirm that there is no potential conflict of interest to disclose in relation to this article.
Informed Consent
Completed written informed consent was obtained.
Author Contributions
Conceptualization: AB, AT, EO and VP; methodology: AB, AT, YR and EO; software: AS, ND; validation: TI, LV and IM; formal analyses: LV, AS and ND; investigation: EO, MB, AK, DG and AK; data curation: YR, EO, AS, ND, LV, TI, IM, MB, AK, DG and AK; writing - original draft preparation: AB, YN and EO; writing - review and editing: AB, AT and VP; writing - literacy search: AK, DG and AK; supervision: VP; project administration: VP. All authors have read and agreed to the published version of the manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
Abbreviations
ACE1: angiotensin-converting enzyme type 1; ACE2: angiotensin-converting enzyme type 2; AGT: angiotensinogen; AGTR1: angiotensin II receptor type 1; AGTR2: angiotensin II receptor type 2; CI: confidence interval; CRP: C-reactive protein; CVD: cardiovascular disease; DBP: diastolic blood pressure; DNA: deoxyribonucleic acid; OR: odds ratio; HWE: Hardy-Weinberg equilibrium; RAS: renin-angiotensin system; RR: respiratory rate; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; SBP: systolic blood pressure; SNP: single nucleotide polymorphism; SpO2: average value of oxygen saturation measured with a pulse oximeter
| References | ▴Top |
- Podzolkov VI, Bragina AE, Tarzimanova AI, Vasilyeva LV, Ogibenina ES, Bykova EE, Shvedov II, et al. Arterial hypertension and severe COVID-19 in hospitalized patients: data from a cohort study. Rational Pharmacotherapy in Cardiology. 2023;19(1):4-10.
- Podzolkov VI, Tarzimanova AI, Bragina AE, Shvedov II, Bykova EE, Ivannikov AA, Vasilyeva LV. Damage to the cardiovascular system in patients with SARS-CoV-2 coronavirus infection. Part 1: predictors of the development of an unfavorable prognosis. Rational Pharmacotherapy in Cardiology. 2021;17(6):825-830.
- Tarzimanova AI, Bragina AE, Sokolova EE, Vargina TS, Pokrovskaya AE, Safronova TA, Loriya IZ, et al. Predictors of premature ventricular contractions development in patients with SARS-CoV-2 infection. J Clin Med Res. 2024;16(5):243-250.
doi pubmed pmc - Dessie ZG, Zewotir T. Mortality-related risk factors of COVID-19: a systematic review and meta-analysis of 42 studies and 423,117 patients. BMC Infect Dis. 2021;21(1):855.
doi pubmed pmc - Degarege A, Naveed Z, Kabayundo J, Brett-Major D. Heterogeneity and risk of bias in studies examining risk factors for severe illness and death in COVID-19: A Systematic Review and Meta-Analysis. Pathogens. 2022;11(5):563.
doi pubmed pmc - Sever P, Johnston SL. The Renin-Angiotensin system and SARS-CoV-2 infection: a role for the ACE2 receptor? J Renin Angiotensin Aldosterone Syst. 2020;21(2):1470320320926911.
doi pubmed pmc - Amara A, Mrad M, Sayeh A, Lahideb D, Layouni S, Haggui A, Fekih-Mrissa N, et al. The effect of ACE I/D polymorphisms alone and with concomitant risk factors on coronary artery disease. Clin Appl Thromb Hemost. 2018;24(1):157-163.
doi pubmed pmc - Shahid M, Rehman K, Akash MSH, Suhail S, Kamal S, Imran M, Assiri MA. Genetic polymorphism in angiotensinogen and its association with cardiometabolic diseases. Metabolites. 2022;12(12):1291.
doi pubmed pmc - Li X, Li Q, Wang Y, Li Y, Ye M, Ren J, Wang Z. AGT gene polymorphisms (M235T, T174M) are associated with coronary heart disease in a Chinese population. J Renin Angiotensin Aldosterone Syst. 2013;14(4):354-359.
doi pubmed - Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3' untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81(2):405-413.
doi pubmed pmc - Kobashi G, Hata A, Ohta K, Yamada H, Kato EH, Minakami H, Fujimoto S, et al. A1166C variant of angiotensin II type 1 receptor gene is associated with severe hypertension in pregnancy independently of T235 variant of angiotensinogen gene. J Hum Genet. 2004;49(4):182-186.
doi pubmed - Cafiero C, Rosapepe F, Palmirotta R, Re A, Ottaiano MP, Benincasa G, Perone R, et al. Angiotensin system polymorphisms' in SARS-CoV-2 positive patients: assessment between symptomatic and asymptomatic patients: a pilot study. Pharmgenomics Pers Med. 2021;14:621-629.
doi pubmed pmc - Kouhpayeh HR, Tabasi F, Dehvari M, Naderi M, Bahari G, Khalili T, Clark C, et al. Association between angiotensinogen (AGT), angiotensin-converting enzyme (ACE) and angiotensin-II receptor 1 (AGTR1) polymorphisms and COVID-19 infection in the southeast of Iran: a preliminary case-control study. Transl Med Commun. 2021;6(1):26.
doi pubmed pmc - Jevnikar K, Lapajne L, Petrovic D, Meglic A, Logar M, Vidovic Valentincic N, Globocnik Petrovic M, et al. The role of ACE, ACE2, and AGTR2 polymorphisms in COVID-19 severity and the presence of COVID-19-related retinopathy. Genes (Basel). 2022;13(7):1111.
doi pubmed pmc - Ministry of Health of the Russian Federation. Temporary guidelines: Prevention, diagnosis and treatment of new coronavirus infection (COVID-19). Version 18 (10/26/2023). Available at: https://static-0.minzdrav.gov.ru/system/attachments/attaches/000/064/610/original/ВМР_COVID-19_V18.pdf. Accessed June 04, 2024.
- COVID-19 treatment guidelines panel. Coronavirus disease 2019 (COVID-19) treatment guidelines. National Institutes of Health. Available at https://www.covid19treatmentguidelines.nih.gov/. Accessed June 04, 2024.
- Kuznetsova T, Staessen JA, Thijs L, Kunath C, Olszanecka A, Ryabikov A, Tikhonoff V, et al. Left ventricular mass in relation to genetic variation in angiotensin II receptors, renin system genes, and sodium excretion. Circulation. 2004;110(17):2644-2650.
doi pubmed - Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74.
doi pubmed pmc - https://www.internationalgenome.org.
- Moreno V, Gonzalez J, Pelegri D. SNPassoc: SNPs-based whole genome association studies. R package version 2.1-0. 2022. Available at: https://CRAN.R-project.org/package=SNPassoc. Accessed January 14, 2024.
- Sinnwell JP, Shaid DJ. haplo.stats: Statistical analysis of haplotypes with traits and covariates when linkage phase is ambiguous. R package version 1.9.5. 2023. Available at: https://CRAN.R-project.org/package=haplo.stats. Accessed January 14, 2024.
- Palmirotta R, Barbanti P, Ludovici G, De Marchis ML, Ialongo C, Egeo G, Aurilia C, et al. Association between migraine and ACE gene (insertion/deletion) polymorphism: the BioBIM study. Pharmacogenomics. 2014;15(2):147-155.
doi pubmed - Ghafouri-Fard S, Noroozi R, Omrani MD, Branicki W, Pospiech E, Sayad A, Pyrc K, et al. Angiotensin converting enzyme: A review on expression profile and its association with human disorders with special focus on SARS-CoV-2 infection. Vascul Pharmacol. 2020;130:106680.
doi pubmed pmc - Larsson SC, Mason AM, Back M, Klarin D, Damrauer SM, Million Veteran P, Michaelsson K, et al. Genetic predisposition to smoking in relation to 14 cardiovascular diseases. Eur Heart J. 2020;41(35):3304-3310.
doi pubmed pmc - Kim HK, Lee H, Kwon JT, Kim HJ. A polymorphism in AGT and AGTR1 gene is associated with lead-related high blood pressure. J Renin Angiotensin Aldosterone Syst. 2015;16(4):712-719.
doi pubmed - Zhu M, Yang M, Lin J, Zhu H, Lu Y, Wang B, Xue Y, et al. Association of seven renin angiotensin system gene polymorphisms with restenosis in patients following coronary stenting. J Renin Angiotensin Aldosterone Syst. 2017;18(1):1470320316688774.
doi pubmed pmc - Gemmati D, Tisato V. Genetic hypothesis and pharmacogenetics side of renin-angiotensin-system in COVID-19. Genes (Basel). 2020;11(9):1044.
doi pubmed pmc - Gupta K, Kaur G, Pathak T, Banerjee I. Systematic review and meta-analysis of human genetic variants contributing to COVID-19 susceptibility and severity. Gene. 2022;844:146790.
doi pubmed pmc - Yamamoto N, Ariumi Y, Nishida N, Yamamoto R, Bauer G, Gojobori T, Shimotohno K, et al. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944.
doi pubmed pmc - Liu PP, Blet A, Smyth D, Li H. The science underlying COVID-19: implications for the cardiovascular system. Circulation. 2020;142(1):68-78.
doi pubmed - Caputo I, Caroccia B, Frasson I, Poggio E, Zamberlan S, Morpurgo M, Seccia TM, et al. Angiotensin II promotes SARS-CoV-2 infection via upregulation of ACE2 in human bronchial cells. Int J Mol Sci. 2022;23(9):5125.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Clinical Medicine Research is published by Elmer Press Inc.